近年の大規模なDNA配列決定技術の進展にともない,生物の遺伝情報であるゲノム配列が多くの種で決定されている.カイコガにおいてもゲノムプロジェクトがほぼ完了し,遺伝子配列が網羅的に明らかにされている(Mita et al., 2004; Qiangyou et al., 2004).このような遺伝子情報に基づいて,細胞を可視化する技術として組織中のmRNAを検出するin situ hybridization法,タンパク質を検出する免疫組織化学的手法,そしてトランスジェニック技術と遺伝子プロモーターを利用した分子遺伝学的手法があげられる.
前者2つの手法が組織の固定を必要とし「死んだ」動物でしか適用できないのに対し,分子遺伝学的手法は「生きた」動物の細胞をin vivoで可視化できるという大きな利点がある.さらに,この手法は単にニューロンの可視化にとどまらず,神経伝達の阻害による特定の神経回路の機能解析や機能遺伝子の導入によるニューロンの機能改変をin vivoで行うことを可能にし,遺伝子から行動を「みる」ために必要不可欠な技術となっている.
このような分子遺伝学的手法による脳神経系の解析はこれまで遺伝子操作技術の発達したキイロショウジョウバエ(Drosophila melanogaster)の独壇場であったが,2000年に田村らにより,カイコガでトランスジェニック体の作出法が確立され(Tamura et al., 2000),カイコガ脳神経系の研究に分子生物学的手法を導入することができるようになった.ここではわれわれが進めているカイコガ脳神経系への分子遺伝学的手法の適用について紹介する.
カイコガでは,トランスポゾンの一種であるpiggyBacを利用したトランスジェニック体の作出法が確立されている(Tamura et al., 2000).トランスジェニック体で特定のニューロンで外来遺伝子を発現させるためには,ターゲットとする細胞で発現することが知られている遺伝子のプロモーター領域を利用する(Fig. 2A).ニューロンを可視化するためのレポーター遺伝子としては染色の操作の必要がなく,生物が生きた状態で検出できる緑色蛍光タンパク質(GFP),赤色蛍光タンパク質(DsRed)などの蛍光タンパク質が主に使用される.図?に人工プロモーターである3xP3プロモーターを用いて,カイコ胚と幼虫の単眼の感覚ニューロンをDsRedで可視化した例を示した.これらのレポーター遺伝子は比較的均一に細胞質に分布するため,発現したニューロン全体の形態を可視化することができる.
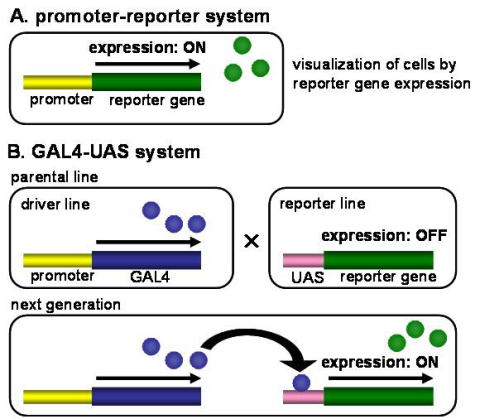
Fig. 2 Schematic diagram of reporter gene expression systems in transgenic silkmoths.
(A) In the promoter-reporter system, promoter sequence is placed immediately upstream of reporter gene and directly activates reporter gene expression. (B) In the GAL4-UAS system, reporter gene expression is activated via GAL4-UAS yeast transcriptional system.
一方で,ショウジョウバエでは細胞内局在シグナル配列を付加した蛍光タンパク質を用いることで,シナプス末端などニューロンの特定の領域の可視化がなされており,カイコガでもそのようなレポーター系統の作出が待たれる.また,レポーター遺伝子にカルシウム感受性蛍光タンパク質G-CaMPを用いることで神経活動のイメージングを行うことができる.同じトランスジェニック系統内の個体は遺伝的に均一であるため,一度トランスジェニック系統を作出すれば,異なる個体で同じ細胞を標識することができる.
プロモーターによる導入遺伝子発現調節の発展型として酵母由来転写因子GAL4とその標的配列であるUASを利用したGAL4-UASシステムがショウジョウバエを中心に広範に使われている.このシステムでは,プロモーター下にGAL4遺伝子配列をつないだ組換え遺伝子を持つ系統とUAS下にリポーター遺伝子をつないだ組み換え遺伝子を持つ系統を別個に作出する.両者を交配して得られた次世代個体ではGAL4がUASに結合し,リポーター遺伝子が発現する(Fig. 7B).GAL4-UASシステムの利点として,発現細胞の特異性を決定するGAL4系統と,レポーター遺伝子を発現するUAS系統が別の系統として分離されていることがあげられる.すなわち,UAS下流に異なる遺伝子をつないだ系統を交配に用いることで,1回の交配実験により簡便にさまざまなレポーター遺伝子やエフェクター遺伝子を同じ細胞で再現的に発現できる.
カイコガにおいても2003年に今村らにより,GAL4-UASシステムが体組織で正常に機能することが報告された(Imamura et al., 2003).そこでわれわれはまず,GAL4-UASシステムが脳神経系で正しく機能することを検証するために,2種類の神経ペプチドホルモンのプロモーター配列の下流にGAL4をもつトランスジェニック系統をそれぞれ作出した.これらの系統とUAS-GFP系統の交配で得られた次世代個体では神経ペプチド分泌細胞で特異的にGFP蛍光が観察された(Yamagata et al., 2008) .この結果は,遺伝子プロモーターを利用してカイコガ脳の特定のニューロンを可視化するためにGAL4-UASシステムが有用であることを示している.
そこで,このシステムを利用してフェロモン情報の触角から脳への空間的な情報入力パターンを調べるために,フェロモン主成分であるボンビコールの受容体遺伝子BmOR114)と副成分であるボンビカールの受容体遺伝子BmOR315)のプロモーター領域下でGAL4を発現する系統を作出した.これまでに,これらの系統とUAS-GFP系統の交配で得られた次世代個体の解析から,触角で受容されたボンビコールとボンビカールの情報が,嗅覚系一次中枢である触角葉の異なる領域に伝達されることを見出している(櫻井ら,未発表).さらに,これらのGAL4系統をUAS-GCaMP系統と交配し,触角をフェロモンで刺激したときの触角葉のカルシウムイメージングに成功している.
このように,カイコガのフェロモン情報処理に関与するニューロン群の構造と機能の理解に分子遺伝学的手法が利用できるようになってきた.今後の課題として,毒素遺伝子などの細胞死や神経伝達を阻害する遺伝子を発現するUAS系統を作出することがあげられる.これにより,可視化したニューロンの機能を阻害し,特定の神経回路がフェロモン源定位行動に果たす役割を個体レベルで知ることができる.さらには,可視化したニューロンに機能遺伝子を導入し,積極的にニューロンの機能を改変することも可能になるだろう.たとえば,イオンチャネルや神経伝達物質受容体遺伝子の導入によりニューロンの特性を遺伝子レベルで改変することで,フェロモン源定位行動の基にある神経回路の機能を改変し,行動を人為的にデザインすることができるようになるかもしれない.
■Genetic engineering techniques
Due to recent advances in large-scale DNA sequencing techniques, genome sequences up to the entire genetic information of organisms have been determined in many species. The silkmoth genome project has almost been completed and the silkmoth genome sequence has been comprehensively elucidated (Mita et al., 2004; Qiangyou et al., 2004). There are several methods to visualize neurons in a region of interest in the nervous system, which are based on the genomic information mentioned above; In situ hybridization method for detecting mRNAs in tissue of interest, immunohistochemical detection methods for specific proteins based on cloned gene products, and genetic engineering methods utilizing transgenic techniques, in particular the use of information on specific promoters and the expression of reporter genes.
In situ hybridization (ISH) is used to visualize the localization of nucleic acids containing specific sequences. The method is usually used for detecting mRNA, permitting a scientist determine when and where a specific gene is expressed in the nervous system. This technique is suitable for identifying which neurons express a gene, although it does not reveal where the functional protein product is localized within the cell. Briefly, a complementary sequence, that would bind to an mRNA sequence of interest by hybridization, i.e. formation of a double-stranded product held together by hydrogen bonds (such as in the DNA), is tagged with some means for visualization, for example a fluorescent compound.
Immunohistochemistry (IHC) is used to visualize the presence of proteins and other biomolecules. This technique depends on antibodies to recognize and bind to specific epitopes (antigens) on proteins or other molecules. For the production of antibodies, the purified antigen is necessary and a good way to obtain purified proteins or peptides in quantity is to clone the gene sequence in question and to have the antigen or a modified protein bearing a wanted epitope produced by an expression system. The antigen can then by purified in large quantities sufficient for immunization of the animals used to produce the antibodies.
In situ hybridization and immunohistochemical methods are only applicable post-mortem because processes such as enzymatic reactions and diffusion barriers that are present in living organisms interfere with these procedures. On the contrary, genetic engineering methods can be used for visualization in living tissues in vivo. Furthermore, these methods can be used to manipulate neural activity of interest.
A reporter gene is a nonendogenous gene encoding an enzyme or fluorescent protein whose expression is controlled by a promoter for a separate (endogenous) gene of interest. Therefore, it is possible to examine the spatial and temporal expression of the gene by measuring the expression of the reporter.
In addition to visualizing neural activity of interest using genetic engineering meth a scientist can also stimulate or inhibit neural activity using genetic engineering methods in combination with optical methods. There are many advantages in using the genetic engineering methods in combination with optical methods of manipulation over electrophysiological or magnetic methods: the genetic engineering methods can be targeted to specific cell types within the neural tissue, large populations of neurons can be simultaneously controlled, and off-target effects from neighboring brain regions are easier to avoid. Both electrical microstimulation and optical stimulation of light-sensitive ion channels, such as channelrhodopsin-2 (ChR2) allow investigators to control neural activity at short time scales. However, optical stimulation has distinct advantages over electrical microstimulation. Stimulation of neurons using light eliminates artifacts that can be caused by electrical stimulation. Also, because light-activated channels are genetically encoded, investigators have the possibility to selectively target specific cell types, enhancing the ability to distinguish the contribution of activity from particular populations of neurons. For example, ChR2 and halorhodopsin (NpHR) allow fast neuronal activation and inactivation, respectively. ChR2 is a cation channel that allows sodium ions to enter the cell upon illumination by ~470 nm blue light, while NpRH is a chloride pump that activates upon illumination by ~580 nm yellow light. Both constructs rapidly activate upon illumination with the appropriate wavelength of light, and rapidly turn off without proper illumination.
The fruit fly Drosophila melanogaster is the insect that is mostly used to analyze the nervous system using the genetic engineering methods because of the long tradition of genetic manipulation techniques in this fly species. Tamura et al. (2000) developed methods for producing transgenic silkmoths, which allow scientists to apply a large grid of genetic engineering methods to the silkmoth nervous system. In the following paragraphs, we briefly describe the examples of how to apply the genetic engineering methods to the silkmoth nervous system.
The process of creating a transgenic silkmoth takes advantage of transposons which are mobile genetic elements that move by way of transition between chromosomes. It usually encodes a specific enzyme called a transposase which acts on a specific DNA sequence at each end of the transposon, causing it to be inserted into a new target DNA site. The production of a transgenic silkmoth is established using the transposon piggyBac (Tamura et al., 2000). The first step in producing a transgenic silkmoth is to use recombinant DNA technology to make a transgenic construct. This construct must contain (1) the DNA sequence encoding the transgene and (2) the necessary promoter sequences for the expression of the gene (Fig. 7). Simply inserting a gene into the genome will not cause it to be expressed: endogenous cellular machinery must recognize a promoter sequence in order to transcribe the DNA into mRNA. Promoters control the expression of genes so that they are expressed in specific cells and tissues at specific time. Next, a plasmid containing a DNA construct is inserted into piggyBac. Then the transgenic construct is injected into a very young silkmoth embryo along with a separate plasmid containing the gene encoding the transposase. If this technique is performed correctly, the injected gene enters the germ line as the result of a transposition event. As reporter genes, we use green fluorescent protein (GFP) or red fluorecent protein (DsRed) to visualize neurons of interest because GFP and DsRed can be detected in living silkmoths. Fig. ? shows an example in which artificial promoter, 3xP3 and DaRed as the reporter gene are used to visualize sensory neurons in a silkmoth embryo and the larval ocelli. These reporters are distributed homogeneously in the cytoplasm so that the neurons expressing the reporters can be visualized. Neuronal activity of interest can be visualized for exampe by using G-Camp as a reporter gene. Because the same transgenic lineage is genetically homogenous the same identified neurons can be labeled once making the transgenic lineage

Fig.7 Schematic diagram of reporter gene expression systems in transgenic silkmots.Schematic diagram of reporter gene expression systems in transgenic silkmots. (A) In the promoter-reporter system, promoter sequence is placed immediately upsteam or reporter gene and directly activates reporter gene expression. (B) In the GAL4/UAS system, reporter gene expression is activated via the GAL4/UAS yeast transcriptional system.
Specific regions of interest in neurons , such as synaptic terminals can be visualized in fruit fly using fluorescent protein provided with an intracellular signal molecule (sequence?). This technique will be applied in the silkmoth in the near future.
The simplest method of making a transgenic animal is to use a single DNA construct, as just desicribed, in which a promoter directly regulates the expression of a transgene. An alternative approach is a binary expression system that uses multiple constructs to further refine the spatial and temporal expression of a transgene. This is typically done by creating two different lines of transgenic animals, each line expressing one of the required constructs, and then mating the animals to produce offspring that express both constructs. These flexible expression systems have numerous advantages. The main advantage is that they can provide control over the timing and location of transgene expression. Another primary advantage is the ability to mix and match combinations of transgenes, saving time and effort in making new transgenic constructs.
The Gal4/UAS system is a binary expression system primarily used in Drosophila, though it has been applied to silkmoth (Imamura et al., 2003). In one construct, a cell-specific promoter drives the expression of the gene encoding Gal4, a transcription factor normally expressed in yeast. In a second construct, a transgene of interest is regulated by a promoter sequence called an upstream activation sequence (UAS). The Gal4 protein binds to the UAS sequence and leads to high expression in cells defined by the promoter regulating Gal4 (Fig.7 B). Imamura et al., (2003) reported that the Gal4/UAS system normally functions in the body tissue (Imamura et al., 2003). In order to verify that the Gal4/UAS system can normally function in the brain tissue of the silkmoth, we made two kinds of transgenic silkmoth, each of which has different neuropeptide promoter sequences on Gal4 upstream region and observed that endocrine cells producing the neuropeptides were labeled by GFP in the F1 that was obtained by crossing the two kinds of transgenic silkmoths with a UAS-GFP line (Yamagata et al., 2008) . This results shows that the Gal4/UAS system is useful to visualize silkmoth brain neurons of interest.
In order to investigate the spatio-temporal pattern of pheromone input from antenna to brain, we made two transgenic silkmoth strains each of which expresses Gal4 under the control of promoters for bombykol receptors, detecting the major component of silkmoth sex pheromone, and the other one for bombykal receptors, detecting the minor pheromone component. Besides, we made an UAS-GFP line separately. Then we obtained the F1 from crosses of each of the GAL4 lines and the UAS-GFP line to confirm that information of bombykol and bombykal are transmitted to different regions in the antennal lobe, the first olfactory center in the brain (Sakurai et al., unpublished). In addition, we succeed in calcium imaging of antennal lobe input activity in response to pheromone stimulation using the F1s of the above Gal4 lines and a UAS-GCaMP line.
Now that genetic engineering is available in the silkmoth, we can hope to understand the structures and functions of neurons related to the pheromone information processing in the silkmoth. We aim to make UAS systems expressing genes to induce cell death or to inhibit neural transmission by toxic gene products. Using these UAS systems, we can infer the functional role of neuronal circuits of interest in pheromone orientation behavior at individual level and even change the function of individual neurons. For example, changing the neuronal characteristics at genetic level by introducing ion channels and/or ligand-gated channels into neurons would allow us to change the neuronal circuit underling the pheromone orientation behavior and even to altersilkmoth behavior artificially in well-controlled ways.